Taxonomic data-mining: converting printed dichotomous keys to digital pathway keys
Ever since the mid-18th century, published, hard copy, dichotomous or pathway keys were the main tools used to describe and provide guides for identifying animal and plant taxa.
Since many of these published keys are out-of-print or regarded as too difficult for non-taxonomists to use, they are likely to be overlooked or ignored.
To illustrate this point, consider one page (of this 50-page key) of the very detailed key shown in Figure 1, which is part of a major, out-of-print, published key for identifying over 800 insects and spiders found in Asian rice crops.

Notice that line drawings have been referenced to illustrated certain morphological features, such as Fig. 406 and Fig 407 for couplet branch 203 and Fig. 408 for couplet branch 204. When using the published key, you needed to turn to page 88 to see these figures, as shown of Figure 2 below.
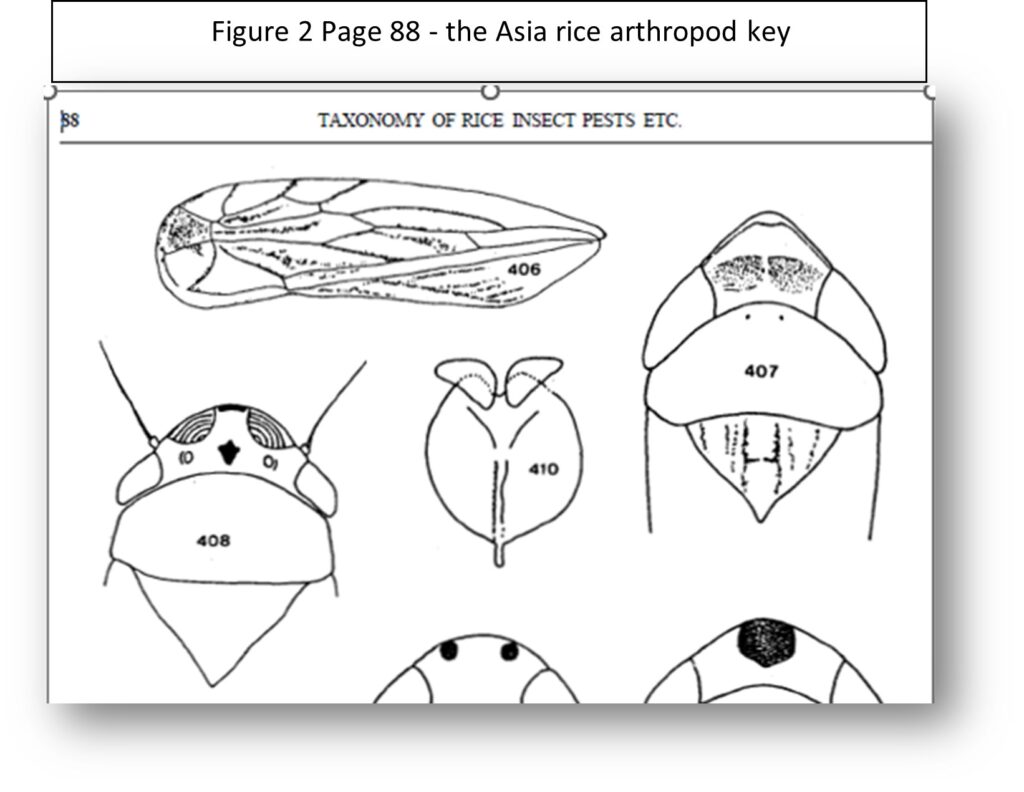
Since identifying pest insects and predatory and parasitic insects and spiders can be very important in managing pest problems in rice, the huge effort involved in collecting, determining the taxonomy of these arthropods, and developing the original published keys, was well worth it.
To make such information more easily accessible, the Lucid pathway key facility has been recently incorporated into Lucid version 4.
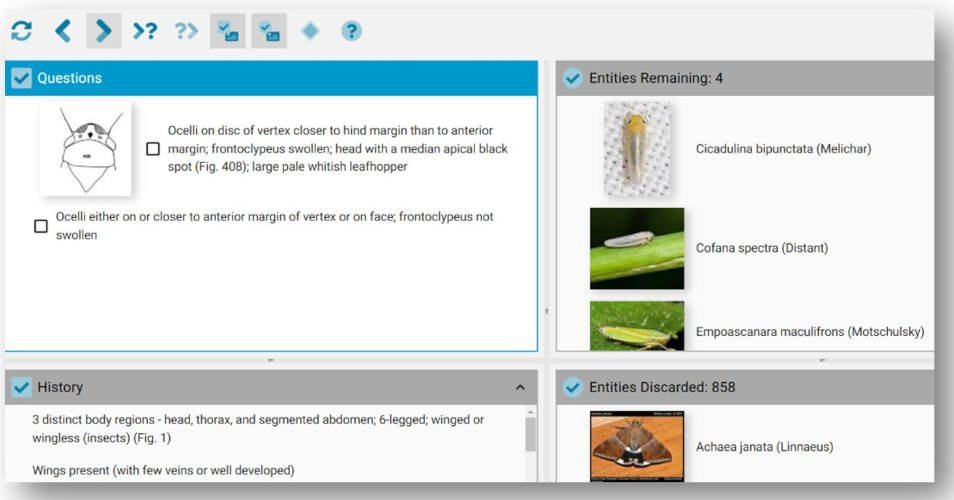
Figure 3 (below) shows how the Lucid Player presents this specific key. In this case the top two windows show the current Couplet Question and the Entities Remaining, while the bottom two windows show the selection History and Entities Discarded.
Figure 3. Screen shot of couplet 204 in the Asian rice arthropod Lucid pathway key.
You can read more about this Lucid pathway key and a similar key for West African rice arthropods at New interactive pathway keys for identifying insect pests of rice and their natural enemies - IAPPS (plantprotection.org), showing how additional online support can be further provided by the use of images and glossaries. |